Microfluidics Cell Assay Chips
Miniaturized biochemical analysis system can be applied to autonomous instrument for environmental field measurement, automatic chemical analyzer and biomedical field such as blood analysis, micro dosing system, DNA analysis and etc. In our lab, we focus on high-throughput single-cell assays to characterize individual cell properties.
|
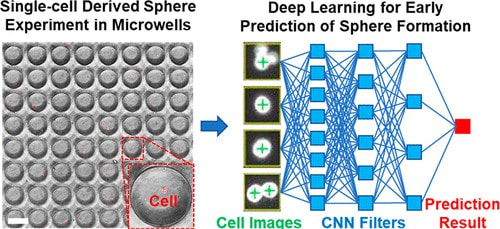
Early Prediction of Single-Cell Derived Sphere Formation Rate Using Convolutional Neural Network Image Analysis
Functional identification of cancer stem-like cells (CSCs) is an established method to identify and study this cancer subpopulation critical for cancer progression and metastasis. The method is based on the unique capability of single CSCs to survive and grow to tumorspheres in harsh suspension culture environment. Recent advances in microfluidic technology have enabled isolating and culturing thousands of single cells on a chip. However, tumorsphere assay takes a relatively long period of time, limiting the throughput of this assay. In this work, we incorporated machine learning with single-cell analysis to expedite tumorsphere assay. We collected 1,710 single-cell events as the database and trained a convolutional neural network model that predicts whether a single cell could grow to a tumorsphere on Day 14 based on its Day 4 image. With this future-telling model, we precisely estimated the sphere formation rate of SUM159 breast cancer cells to be 17.8% based on Day 4 images. The estimation was close to the ground truth of 17.6% on Day 14. The preliminary work demonstrates not only the feasibility to significantly accelerate tumorsphere assay but also a synergistic combination between single-cell analysis with machine learning, which can be applied to many other biomedical applications. [Yu-Chih Chen, Zhixiong Zhang, and Euisik Yoon, “Early Prediction of Single-cell Derived Sphere Formation Using Convolutional Neural Network Image Analysis,” Analytical Chemistry, 92, 11, 7717–7724, 2020.] [Link] |
|
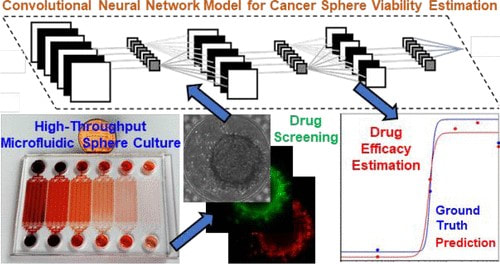
Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis
Despite recent advances in cancer treatment, developing better therapeutic reagents remains an essential task for oncologists. To accurately characterize drug efficacy, 3D cell culture holds great promise as opposed to conventional 2D monolayer culture. Due to the advantages of cell manipulation in high-throughput, various microfluidic platforms have been developed for drug screening with 3D models. However, the dissemination of microfluidic technology is overall slow, and one missing part is fast and low-cost assay readout. In this work, we developed a microfluidic chip forming 1920 tumor spheres for drug testing, and the platform is supported by automatic image collection and cropping for analysis. Using conventional LIVE/DEAD staining as the ground truth of sphere viability, we trained a convolutional neural network to estimate sphere viability based on its bright-field image. The estimated sphere viability was highly correlated with the ground truth (R-value > 0.84). In this manner, we precisely estimated drug efficacy of three chemotherapy drugs, doxorubicin, oxaliplatin, and irinotecan. We also cross-validated the trained networks of doxorubicin and oxaliplatin and found common bright-field morphological features indicating sphere viability. The discovery suggests the potential to train a generic network using some representative drugs and apply it to many different drugs in large-scale screening. The bright-field estimation of sphere viability saves LIVE/DEAD staining reagent cost and fluorescence imaging time. More importantly, the presented method allows viability estimation in a label-free and nondestructive manner. In short, with image processing and machine learning, the presented method provides a fast, low-cost, and label-free method to assess tumor sphere viability for large-scale drug screening in microfluidics. [Zhixiong Zhang, Lili Chen, Yimin Wang, Tiantian Zhang, Yu-Chih Chen*, and Euisik Yoon*, "Label-free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis," Analytical Chemistry, 91, 21, 14093-14100, 2019.] [Link] |
|
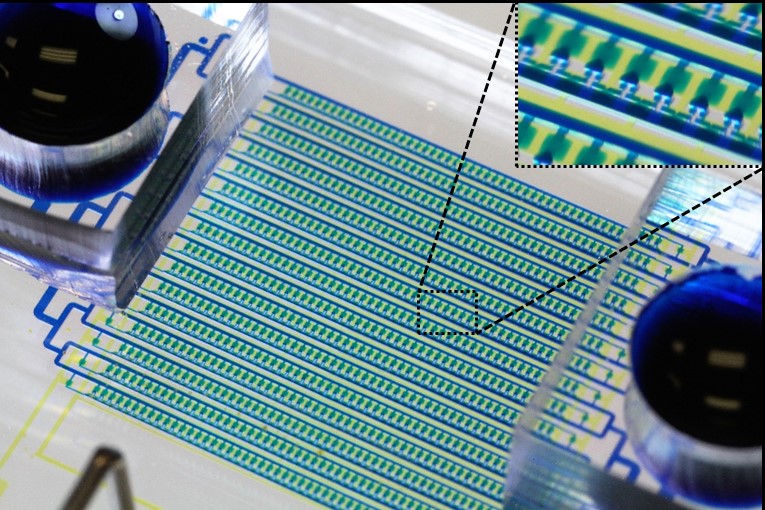
Hydro-Seq Enables Contamination-free High-throughput Single-cell RNA-sequencing for Circulating Tumor Cells
Analyzing individual CTCs as a liquid biopsy has high potential for diagnosis, assessment of prognosis, and treatment in cancer, yet blood samples contain merely tens of cancer cells mixed with millions of WBCs and billions of RBCs. Though investigators have developed various methods to enrich CTCs in blood sample, the challenges of analyzing small number of cells amidst large numbers of residual contaminating cells are still far beyond capabilities of existing tools. We developed a hydrodynamic microfluidic platform (HydroSeq) that removes contaminating blood cells while isolating ~70% of CTCs for single-cell RNA sequencing. We have processed 21 blood samples from breast cancer patients and successfully sequenced 666 CTCs, which are one order of magnitude more than previous studies. Thanks to the large amount of data we acquired, we uncovered the heterogeneous nature of CTCs. Remarkably, different CTC populations (e.g. HER2- epithelial-to-mesenchymal transition (EMT)-like and HER2+ MET-like) can co-exist within a single patient sample. The observed heterogeneity suggests specific targets for therapy that would not be captured by sequencing many CTCs in bulk or with WBC/RBC contamination. The capability to analyze a large number of single CTCs comprehensively advances disease monitoring to the next level. [Yu-Heng Cheng*, Yu-Chih Chen*, Eric Lin*, Riley Brien, Yu-Ting Chen, Zhijian Hao, Woncheol Lee, Hyun Min Kang, Jason Cong, Monika Burness, Sunitha Nagrath, Max Wicha, and Euisik Yoon, “Hydro-Seq Enables Contamination-free High-throughput Single-cell RNA-sequencing for Circulating Tumor Cells,” Nature Communications, 10, 2163, 2019.] [Link] |
|
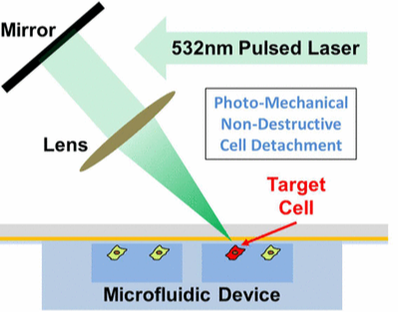
Selective Photomechanical Detachment and Retrieval of Divided Sister Cells from Enclosed Microfluidics for Downstream Analyses
Considerable evidence suggests that self-renewal and differentiation of cancer stem-like cells, a key cell population in tumorgenesis, can determine the outcome of disease. Though the development of microfluidics has enhanced the study of cellular lineage, it remains challenging to retrieve sister cells separately inside enclosed microfluidics for further analyses. In this work, we developed a photomechanical method to selectively detach and reliably retrieve target cells from enclosed microfluidic chambers. Cells cultured on carbon nanotube–polydimethylsiloxane composite surfaces can be detached using shear force induced through irradiation of a nanosecond-pulsed laser. This retrieval process has been verified to preserve cell viability, membrane proteins, and mRNA expression levels. Using the presented method, we have successfully performed 96-plex single-cell transcriptome analysis on sister cells in order to identify the genes altered during self-renewal and differentiation, demonstrating phenomenal resolution in the study of cellular lineage. [Yu-Chih Chen, Hyoung Won Baac, Kyu-Tae Lee, Shamileh Fouladdel, Kendall Teichert, Jong G. Ok, Yu-Heng Cheng, Patrick N. Ingram, A. John Hart, Ebrahim Azizi, L. Jay Guo, Max S. Wicha, and Euisik Yoon, "Selective Photo-Mechanical Detachment and Retrieval of Divided Sister Cells from Enclosed Microfluidics for Downstream Analysis" ACS Nano in Press 2017.] [Link] |
|
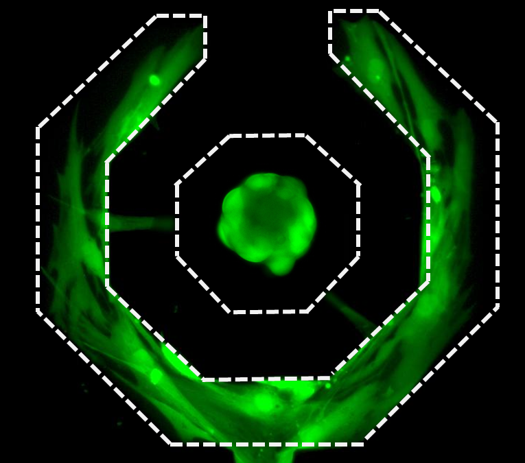
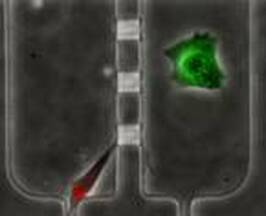
Single Cell Dual Adherent-Suspension Co-Culture Micro-Environment for Studying Tumor-Stormal Interactions with Functionally Selected Cancer Stem-like Cells
In this work, we present a dual adherent/suspension co-culture device, which combines a suspension environment for single-cell tumorsphere assays and an adherent environment for co-culturing stromal cells in close proximity by selectively patterning polyHEMA in indented microwells. By minimizing dead volume and improving cell capture efficiency, the presented platform allows for the use of small numbers of cells (<100 cells). As a proof of concept, we co-cultured single T47D (breast cancer) cells and primary cancer associated fibroblasts (CAF) on-chip for 14 days to monitor sphere formation and growth. Compared to mono-culture, co-cultured T47D have higher tumorigenic potential (sphere formation rate) and proliferation rates (larger sphere size). Furthermore, 96-multiplexed single-cell transcriptome analyses were performed to compare the gene expression of co-cultured and mono-cultured T47D cells. Phenotypic changes observed in co-culture correlated with expression changes in genes associated with proliferation, apoptotic suppression, tumorigenicity and even epithelial-to-mesechymal transition. Combining the presented platform with single cell transcriptome analysis, we successfully identified functional CSCs and investigated the phenotypic and transcriptome effects induced by tumor–stromal interactions. [Yu-Chih Chen, Zhixiong Zhang, Shamileh Fouladdel, Yadwinder Deol, Patrick N. Ingram, Sean P. McDermott, Ebrahim Azizi, Max S. Wicha, and Euisik Yoon, “Single Cell Dual Adherent-Suspension Co-Culture Micro-Environment for Studying Tumor-Stormal Interactions with Functionally Selected Cancer Stem-like Cells” Lab Chip, 2016, 16, 2935-2945.] [Link] |
|
Single Cell Proteolytic Assays to Investigate Clonal Heterogeneity and Cell Dynamics Using an Efficient Cell Loading Scheme
In this work, we developed a microfluidic platform that provides high-efficiency cell loading and simple valveless isolation, so the proteolytic activity of a small sample (10-100 cells) can be easily characterized. Combined with a single cell derived (clonal) sphere formation platform, we have successfully demonstrated the importance of microenvironmental cues for proteolytic activity and also investigated the difference between clones. Furthermore, the platform allows monitoring single cells at multiple time points, unveiling different cancer cell line dynamics in proteolytic activity. The presented tool facilitates single cell proteolytic analysis using small samples, and our findings illuminate the heterogeneous and dynamic nature of proteolytic activity. [Yu-Chih Chen, Yu-Heng Cheng, Patrick Ingram, and Euisik Yoon, “Single Cell Proteolytic Assays to Investigate Clonal Heterogeneity and Cell Dynamics Using an Efficient Cell Loading Scheme,” Scientific Reports, 6, 27154.] [Link] |
|
High-Throughput Single-Cell Derived Sphere Formation for Cancer Stem-Like Cell Identification and Analysis
In this work, we present a 1,024-microchamber microfluidic platform for single-cell derived sphere formation. Utilizing a hydrodynamic capturing scheme, more than 70% of the microchambers capture only one cell, allowing for monitoring of sphere formation from heterogeneous cancer cell populations for identification of CSCs. Single-cell derived spheres can be retrieved and dissociated for single-cell analysis using a custom 96-gene panel to probe heterogeneity within the clonal CSC spheres. This microfluidic platform provides reliable and high-throughput sphere formation for CSC identification and downstream clonal analysis. [Y.-C. Chen, P. N. Ingram, S. Fouladdel, S. P. McDermott, A. Ebrahim, M. S. Wicha and E. Yoon, “High-Throughput Single-Cell Derived Sphere Formation for Cancer Stem-Like Cell Identification and Analysis,” Scientific Reports, 6, 27301. 2016.] [Link] |
|
Paired Single Cell Co-culture Microenvironment Isolated by Two-phase Flow
In this work, two-phase flow was used as the simple isolation method, separating the microenvironments of each individual chamber. As nutrients in an isolated chamber are consumed by cells, media exchange is required. To connect cell culture chamber to the media exchange layer, we demonstrated a 3D microsystem integration technique using vertical connections fabricated by deep reactive-ion etching (DRIE). Utilizing these innovations, we demonstrated co-culture of head and neck squamous cells and endothelial cells to recapitulate tumor proliferation enhancement in the vascular endothelial niche. [Yu-Chih Chen, Yu-Heng Cheng, Hong Sun Kim, Patrick N. Ingram, Jacques E. Nor and Euisik Yoon, “Paired single cell co-culture microenvironments isolated by two-phase flow with continuous nutrient renewal,” Lab Chip, 2014, 14, 2941-2947.] [Link] |
|
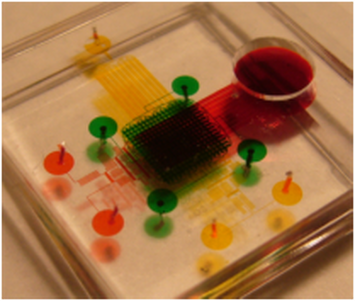
High-throughput PDT Screening Chip
In vitro efficacy assay is a routine prescreen step for anti cancer drug development including various showing up photosensitizers (drugs used in photodynamic therapy or PDT). As photodynamic therapy’s action mechanism involves essential interactions among photosensitizer, oxygen and light, an in vitro test of photosensitizer requires controls of all three factors, and more detailed preclinical tests could provide better guidance for its potential optimum in-vivo protocols. [X. Lou, G. Kim, H. Yoon, Y. Koo, R. Kopelman, E. Yoon, "A High-Throughput Photodynamic Therapy Screening Platform with On-Chip Control of Multiple Microenvironmental Factors", Lab Chip, vol. 14, pp. 892-901, 2014.] [X. Lou, G. Kim, Koo Lee, R. Kopelman, and E. Yoon. “Investigating Photodynamic Efficiency of Tumor Targeted Nanoparticular Photosensitizer Using Microfluidic Chips,” International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS ’11), 2011, pp. 2058-2060] [Link] Video Clip: https://www.youtube.com/watch?v=avCLW_2EZKQ News Cover: http://www.eecs.umich.edu/eecs/about/articles/2014/biochips-for-better-cancer-therapy.html |
|
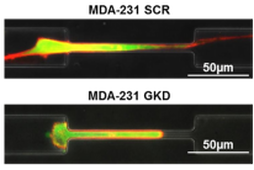
Single Cell Migration Chip
Cell migration is an essential process in angiogenesis, cancer metastasis, wound healing, inflammation and even embryogenesis. With this mechanism, cells can move into proper positions and enact appropriate functions in the body. There are a number of previous works reported to study cell migration in a microfluidic chip. Most works, however, have loaded and cultured thousands of cells in the same channel; thus, it is impossible to trace heterogeneous cell behaviors. In this work, we develop a high capture rate cell migration chip capable of tracing cell migration at a single-cell resolution readily after cell loading. [Y.-C. Chen, X. Lou, P. Ingram, and E. Yoon, “Single cell Migration Chip Using Hydrodynamic Cell Positioning,” International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS ’11), pp. 1409-1411, Seattle, October 2011.] |
|
PDMS Hydrophobic Surfaces for High-throughput Cancer Spheroid Culture
Hydrophobic surfaces have become popular for anti-biofouling applications, but they can also be attractive platforms for non-adherent mammalian cell culture. Suspension growth of cancer cells allows for the identification and subsequent enrichment of cancer stem cells (CSCs) by confirming their ability to form spheres in suspension. We have achieved successful non-adherent spheroid culture of cancer cells on hydrophobic surfaces, formed in polydimethylsiloxane (PDMS), to be used in CSC enrichment and isolation. These surfaces have additionally been integrated within a single cell capture microfluidic device for high throughput performance. [P. Ingram, M. Im, S. McDermott, M. Wicha, and E. Yoon. “Spheroid Cell Culture on PDMS Hydrophobic Surfaces and Integration into Microfluidic Devices,” International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS ’11), 2011, pp. 1539-1541.] |
|
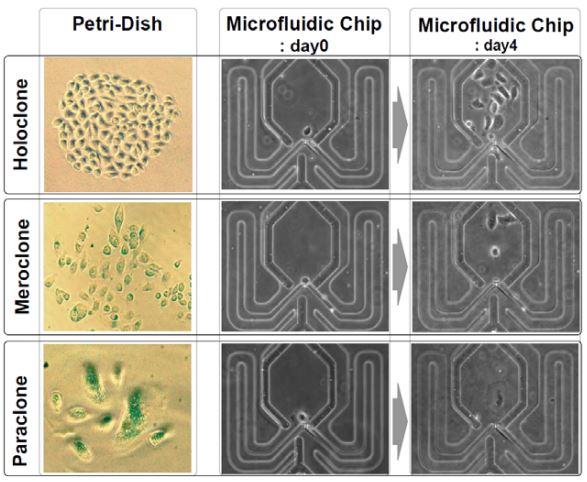
Clonal culture and chemodrug assay of heterogeneous cells (pc3 prostate carcinoma cells) using microfluidic single cell array chips
A major obstacle in cancer research and treatment is that cancer cells often mutate or differentiate into multiple different cell types during experiments. Even testing a drug in a petridish culture might only give information that is averaged across multiple subtypes with very different resistance mechanisms. Recently it was reported that even PC3 human prostate carcinoma cells, a cell line, give rise to a mixture of three clonal phenotypes: holoclones, meroclones, and paraclones. We report a microfluidic array chip that can separate PC3 cancer cells into single cells in each microwell, grow them separately side by side into small uniform clonal colonies and test drugs on them. Using this chip we can detect how many subtypes of clones are present and to see if they respond differently to cancer drug treatments. [J. Chung, P. N. Ingram, T. Bersano-Begey, and E. Yoon, “Traceable clonal culture and chemodrug assay of heterogeneous prostate carcinoma PC3 cells in microfluidic single cell array chips,” Biomicrofluidics, 8, 064103, 2014.] [J. Chung, T. Bersano-Begey, K. Pienta, E. Yoon, “Clonal Culture and Chemodrug Assay of Heterogeneous Cells (PC3 Prostate Carcinoma Cells) Using Microfluidic Single Cell Array Chips,” International Conference on Miniaturized Systems for Chemistry and Life Sciences (mTAS’09), pp. 21-23, November 2009.] |
|
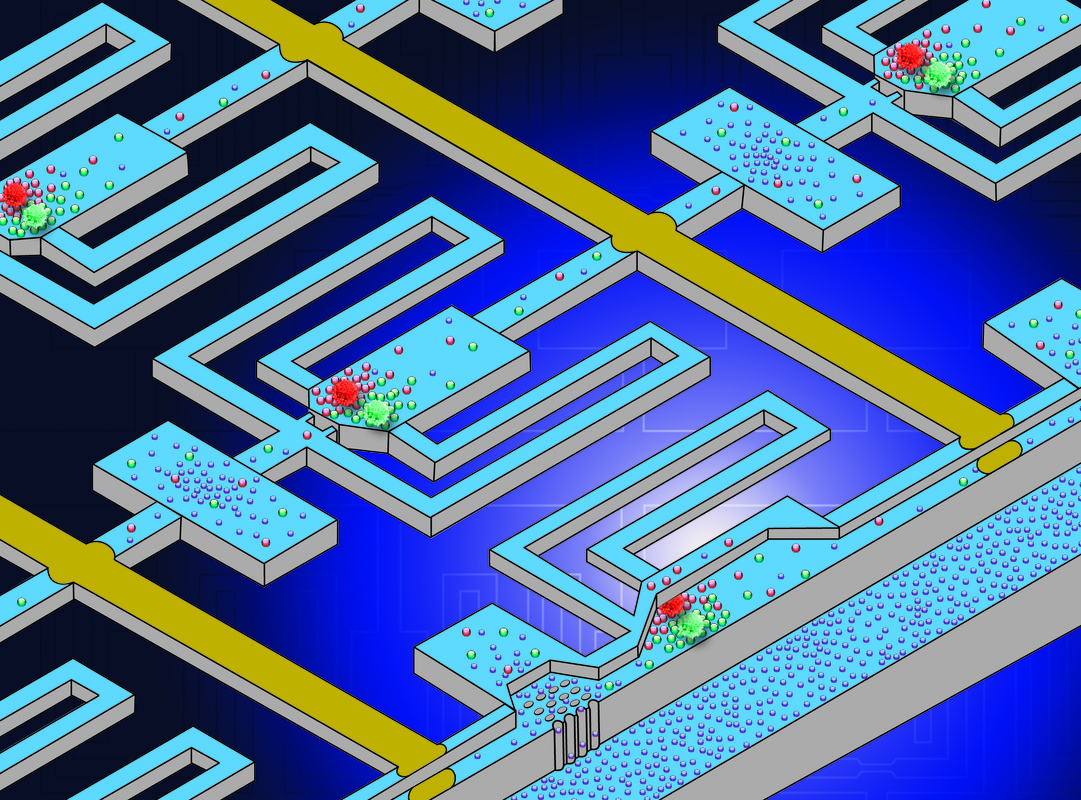
Microfluidic array chip for analysis of pairwise cell interaction by temporal stimulation of secreted factors using chamber isolation
Cellular processes like cancer metastasis and stem cell proliferation and differentiation are highly susceptible to cell-to-cell interaction. Cell-cell interaction analysis has been used to elucidate many cellular processes, but conventional methods are limited to averaged measurement of hundreds or even millions of cells, which is not always desirable and makes high-throughput testing problematic. To address this issue, we propose a new microfluidic device which can analyze pairwise cell interactions in isolated microchamber arrays, enabling hundreds of simultaneous tests and potential high-throughput screening of useful cell pairings. Soluble factors secreted by cells in each pair can quickly reach high physiological concentrations when trapped in a small chamber in proximity to each other, rather than being diluted in larger amounts of media as in conventional methods. Media exchange for cell viability is achieved by quick washes and media perfusion alternated with media isolation phases to expose cells to accumulated signals. [Y.-J. Kim, T. Bersano-Begey, X. Lou, J. Chung, and E. Yoon, “Microfluidic Array Chip for Analysis of Pairwise Cell Interaction by Temporal Stimulation of Secreted Factors Using Chamber Isolation,” International Conference on Miniaturized Systems for Chemistry and Life Sciences (mTAS’09), pp. 1772-1774, November 2009] |
|
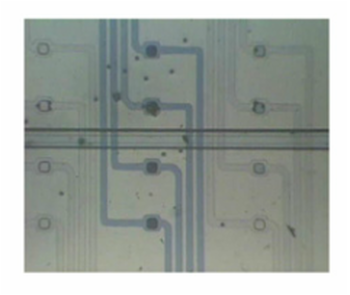
Microfluidic Array Chip for Single-Cell Assay
There has been growing interest in single-cell assays to understand single-cell behavior more accurately. To address this, different methods for single-cell assay using microfluidics have been developed by various research groups. Our research however developed the first microfluidic array platform which allows to both capture and isolate single-cells, and to monitor cell behavior in different environments. [Y.-J. Kim, J. Chung, H.-K Lee and E. Yoon, “Microfluidic Array Chip For Sin-gle-Cell Isolation Using Two-Way Pneumatic Actuation,” IEEE International Conference on MEMS, Technical Digest, pp. 14-17, (2008).] |
|
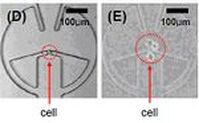
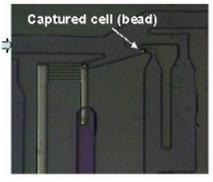
Highly Efficient Single Cell Capturing Using Hydrodynamic Guiding Structures
A microfluidic chip array for high-throughput single cell assay with an efficient cell capture rate was presented. The single cell capture structure utilizes hydrodynamic resistance difference in flow paths and its feasibility was tested. More than 40% of all the injected microbeads were captured in the designated capture sites. The capture structure was expanded in an array of microwells formed at the intersection of rows and columns of orthogonal microchannels. By selectively operating integrated pneumatic valves, we successfully demonstrated cell seeding, reagents injection and cell isolation without cross-contamination. [J. Chung, Y.-J. Kim, I.-J. Cho, and E. Yoon, “Highly Efficient Single Cell Capturing in Microwell Array Using Hydrodynamic Guiding Structures,” Micro Total Analysis Systems (mTAS’08), pp. 477-479, Oct. 2008.] |
|
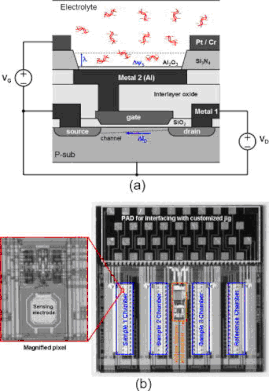
Real-time label-free quantitative monitoring of biomolecules by floating-gate complementary metal-oxide semiconductor sensor
We report a label-free field-effect sensing array integrated with complementary metal-oxide semiconductor (CMOS) readout circuitry to detect the surface potential determined by the negative charge in DNA molecules. For real-time DNA quantification, we have demonstrated the measurements of DNA molecules without immobilizing them on the sensing surface which is composed of an array of floating-gate CMOS transistors. This nonimmobilizing technique allows the continuous monitoring of the amount of charged molecules by injecting DNA solutions sequentially. We have carried out the real-time quantitative measurement of 19 bp oligonucleotides and analyzed its sensitivity as a function of pH in buffer solutions. [S.-J. Kim, K. Yoo, J. Shim, W. Chung, C. Ko, M. Im, L.-S. Kim, and E. Yoon, “Real-time label-free quantitative monitoring of biomolecules without surface binding by floating-gate complementary metal-oxide semiconductor sensor array integrated with readout circuitry,” Applied Physics Letters, Vol. 91, No. 20, pp. 203903-1-203903-3, Nov. 2007.] |
|
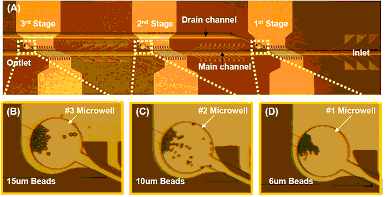
Programmable Magnetic Cell Size Sorter
A programmable magnetic cell sorter to seperate different-sized cells was presented. The cell sorter consists of a series of seperating units, comprising ferromagnetic nickel lines and current-carrying gold lines. The magnetized nickel lines and the current of gold lines can generate local field and attract the cells coated with magnetic nanoparticles. By changing the current, we could control the local magnetic field, which enables the device to sort the target cell sizes selectively. We successfully demonstrated this scheme by sorting three different sizes (6, 10, and 15 .m in diameter) of magnetic beads under the different current of 21, 37, and 68mA/line to the gold lines in the fabricated microfluidic chip. [J. Chung, H.-K. Lee, Y.-J. Kim and E. Yoon, “Programmable Magnetic Cell Size Sorter” Proceedings of microTAS 2007, pp. 793-795, Oct., Paris, France, 2007, pp. 793-795, Oct., 2007.] |
|
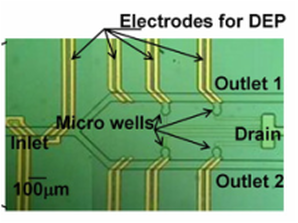
Active cell positioning using Dielectrophoresis (DEP)
Many researches have been performed for the positioning and capturing of cells at single cell level in microfluidic systems for biological and biochemical analysis. However, in most of the previous works it was difficult to expand them into a 2-dimensional array because they only passively captured the cells and did not employ any accurate control of positioning/capturing cells one by one actively. In this work, we propose a new scheme for active positioning control of single cell using dielectrophoresis (DEP). [Byoung-Gyun Kim, Kwang-Seok Yun, and Euisik Yoon, “Active Positioning Control of Single Cell/Microbead in a Micro-Well Array Chip by Dielectrophoresis,” IEEE International Conference on MEMS, Technical Digest pp.702-705, Miami Beach, USA, 30 Jan. – 3 Feb. 2005.] |
|
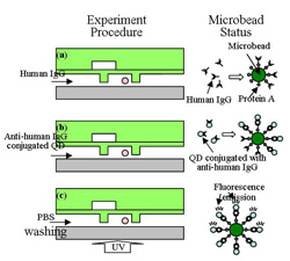
Antibody detection by using single bead-based assay
QD-based detection has attracted research attention because of its numerous advantages such as high sensitivity, narrow emission bandwidth, long-term photostability and tunable multi-color imaging compared to conventional organic dyes. We propose a microbead-based assay for adaptive antibody detection by using a microfluidic chip designed to manipulate single microbead. [K.-S. Yun, D. Lee, H.-S. Kim and E. Yoon, “A Microfluidic Chip for Measurement of Bio Molecules Using Microbead-Based Quantum Dot Fluorescence Assay,” Meas. Sci. Technol. 17, pp. 3178-3183, 2006.] |
|
High throughput cell analysis and drug screening
The need for high-throughput cell analysis/drug screening has been increased with the progress in biotechnology and combinatorial chemistry. In order to address the requirements of expensive equipments and long/tedious experimental works for highthroughput screening, a chip-based cell assay has been developed and reported in recent researches. However, in these previous devices it was relatively difficult to expand them into a two dimensional array with a large number of analysis sites. Also, the flow of cell suspension media or reagents should be maintained to be flowing into the captured cell in order to uphold the cells in each capturing site during the cell culturing and analysis. In this work, we propose a microfluidic chip with multiple micro-wells in two dimension arrays. The proposed chip provides a physical isolation in the captured cells using a PDMS membrane cover, so that a specific reagent can be introduced into each micro-well without continuously maintaining a cell suspension media flow. [Kwang-Seok Yun, and Euisik Yoon, “A Micro/Nano-Fluidic Chip-Based Micro-Well Array for High-Throughput Cell Analysis and Drug Screening,” Proceedings of International Conference on Miniaturized Chemical and Biochemical Analysis Systems (micro TAS’03), pp.861-864, Squaw Valley, California, USA, 5-9 October 2003.] |
|
Single-Cell analysis chip
Cells are usually influenced by a mixture of hormones, ions, and neurotransmitters released from neighboring cells. Dynamic monitoring of a single cell is important for individual cell analysis without the influence of neighboring cells in independently controlled environments. In our research, we have developed a microfluidic chip in which a single-cell is autonomously captured in a cell-positioning site by a pre-defined fluid stream and a specific liquid flow including drugs or reagents can be consecutively supplied to each single-cell through a flow injection channel. [K.-S. Yun, S.-I. Lee, G. M. Lee, and E. Yoon, “Design and fabrication of micro/nano-fluidic chip performing single-cell positioning and nanoliter drug injection for single-cell analysis,” Proceedings of the Micro TAS Symposium (micro TAS’02), Nara, Japan, November 3-7, 2002, pp. 652-654.] |
|
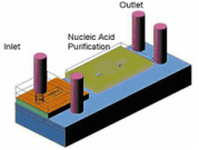
A Disposable DNA sample preparation microfluidic chip
The Nucleic Acid (NA) probe assays typically include PCR to amplify the number of copies of DNA to a detectable level. The PCR technique requires a relatively pure DNA sample in aqueous solution, free of inhibitors during the PCR process. Therefore, the extraction and purification of nucleic acids from biological samples are the critical steps that should be carefully handled in the NA assays. In this work we have proposed a new microfluidic chip to address this sample preparation process for NA probe assays. [Joon-Ho Kim, Byoung-Gyun Kim, Hyukjun Nam, Dae-Eun Park, Kwang-Seok Yun, Jun-Bo Yoon, Jichang You and Euisik Yoon, “A Disposable DNA Sample Preparation Microfluidic Chip for Nucleic Acid Probe Assay”, IEEE International MEMS Conference 2002, Las Vegas, USA, Jan., 2002.] |